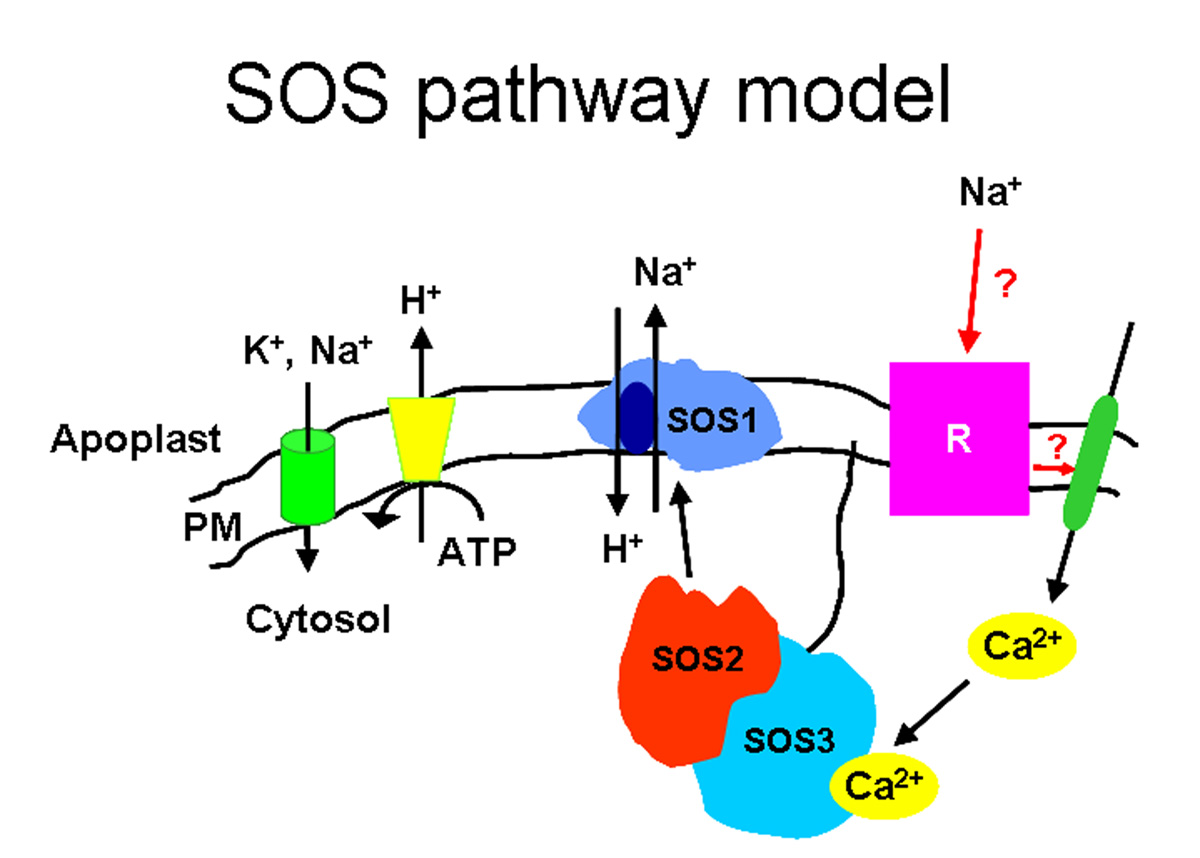
In a genetic screen designed to identify components of the mechanisms controlling salt tolerance in Arabidopsis, several SOSgenes wereid Nine SOS3-like calcium sensor/binding proteins (also identified as AtCBL calcium-binding proteins) have been identified in Arabidopsis (12). Like SOS3, these calcium-binding proteins have no apparent enzymatic activity by themselves and thus are sensor relays (20). The CBL proteins are predicted to possess three to four typical EF-hand calcium-binding motifs flanked by E and F helices (16). Several calcium-binding proteins, including SOS3/CBL4 and CBL1, are associated with membrane fractions (10, 16); this membrane localization is consistent with the idea that many calcium signaling events are initiated by calcium fluxes across membranes (19). At least three calcium-binding proteins, SOS3/CBL4, CBL5 and CBL1, contain a conserved N-myristoylation motif (10). It is possible that this co-translational modification may help tether these calcium sensors to specific membrane patches where calcium flux takes place and/or where target proteins are localized or may contribute to pathway regulation by a calcium-myristoyl switch (10). Members of the CBL family have been implicated in calcium-dependent responses to environmental signals when the plant experiences abiotic stress (3, 7, 14). With funding from the Energy Biosciences Program at the Department of Energy, we are working to add to our understanding of the functions of this important family of calcium-binding proteins. To do this, we are determining: (i) the extent and nature of CBL interactions with other proteins (in vitro and in vivo interaction assays), (ii) the developmental processes regulated by the CBL proteins (phenotypic analysis of loss-of-function mutants, localization of CBL gene activities and protein accumulation) and (iii) if CBL interactions result in additional functions during calcium signaling. The CBL proteins share sequence similarity to the regulatory B subunit of calcineurin. Calcineurin has been identified in organisms from yeast to mammals as a heterodimer with a calcium-binding regulatory subunit (Calcineurin B) and a catalytic calcium/calmodulin-regulated protein phosphatase subunit (Calcineurin A). In Arabidopsis, a Calcineurin A-like 2B-type protein phosphatase has not been identified; instead, a family of SNF1-like serine/threonine protein kinases (SOS2-like or Calcineurin B-like Interacting Protein Kinases, CIPKs) has been identified as calcium-dependent targets for the CBLs (5, 8, 11, 21), interacting through a FISL or NAF domain in the CIPK protein (1, 6, 7). Each CBL protein may interact with several CIPK proteins and certain CIPK proteins can interact with several CBLs. Common interactions between CBLs and CIPKs may allow cross-talk between signaling cascades while preferential associations between CBLs and CIPKs may give rise to specific signaling cascades. The Arabidopsis genome contains twenty-five CIPKs (12) which can be divided into two sub-groups based on the presence or absence of introns. To add to our understanding of the functions of this novel family of protein kinases, we are determining: (i) the developmental processes mediated by the CIPK proteins (phenotypic analysis of single and higher order mutants, localization of CIPK gene activities and protein accumulation), (ii) which CBLs regulate CIPK activity (in vitro and in vivo interaction assays), (iii) the substrate targets for CIPK phosphorylation, and (iv) the biochemical mechanisms that regulate CIPK substrate phosphorylation. ________________________________ REFERENCES 1. Albrecht V, Ritz O, Linder S, Harter K, Kudla J. 2001. The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J 20: 1051-63. 2. Bouché N, Yellin A, Snedden WA, Fromm H. 2005. Plant-specific calmodulin-binding proteins. Annu Rev Plant Biol 56: 435-66. 3. Cheong YH, Kim K-N, Pandey GK, Gupta R, Grant JJ, Luan S. 2003. CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15, 1833-45. 4. Day IS, Reddy VS, Ali GS, Reddy ASN. 2002. Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol 3: 1-24. 5. Gong D, Guo Y, Schumaker KS, Zhu J-K. 2004. The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol 134: 919-26. 6. Guo Y, Halfter Y, Ishitani M, Zhu J-K. 2001. Molecular characterization of functional domains in the protein kinase SOS2 that is required for salt tolerance. Plant Cell 13: 1383-400. 7. Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu J-K. 2002. A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3: 233-44. 8. Halfter U, Ishitani M, Zhu J-K. 2000. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97: 3735-40. 9. Ikura M. 1996. Calcium binding and conformation response in EF-hand proteins Trends in Biol Sci 21: 14-17. 10. Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu J-K. 2000. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12: 1667-78. 11. Kim K-N, Cheong YH, Gupta R, Luan S. 2000. Interaction specificity of Arabidopsis calcineurin B-like sensors and their target kinases. Plant Physiol 124: 1844-53. 12. Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J. 2004. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134: 43-58. 13. Liu J, Ishitani M, Halfter U, Kim CS, Zhu J-K. 2000. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97: 3730-34. 14. Liu J, Zhu J-K. 1998. A calcium sensor homolog required for plant salt tolerance. Science 280: 1943-45. 15. McCormack E, Braam J. 2003. Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol 159: 585-98. 16. Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W. 2002. Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14 Suppl S389-400. 17. Qiu Q-S, Barkla BJ, Vera-Estrella R, Zhu J-K, Schumaker KS. 2003. Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol 132: 1041-52. 18. Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K. 2002. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99: 8436-41. 19. Rudd J, Franklin-Tong V. 2001. Unraveling response-specificity in Ca2+ signaling pathways in plant cells. New Phytol 151: 7-33. 20. Sanders D, Pelloux J, Brownlee C, Harper JH. 2002. Calcium at the crossroads of signaling. Plant Cell 14 Suppl S401-17. 21. Shi J, Kim K-N, Ritz O, Albrecht B, Gupta R, Harter K, Luan S, Kudla J. 1999. Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11: 2393-405.
home | who we are | research | publications | tools | contact us |