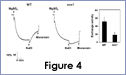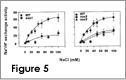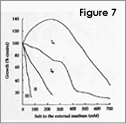
We are interested in understanding how plants control cellular levels of both beneficial and toxic ions during normal growth and development and when the plant experiences an abiotic stress. Our research activities are focused in the following two areas. Abiotic stress - Mechanisms underlying plant adaptation to salt stress . Environmental stresses such as salinity limit crop productivity worldwide. Sodium enters plant cells and inactivates metabolic enzymes leading to cell death and ultimately to death of the plant. Maintaining low levels of sodium in the cell is crucial for plant growth and development. Through comparative studies with various ecotypes of Arabidopsis thaliana (a salt-sensitive plant, a glycophyte) and a related species, Thellungiella halophila (a salt-tolerance plant, a halophyte), our goal is to elucidate the molecular basis of plant salt tolerance. Signal transduction - Mechanisms controlling stimulus-induced changes in intracellular calcium . Mechanisms underlying plant adaptation to salt stress One of the most urgent global problems is finding enough water and land to support the world's food needs. The increasing demand for plant-based agricultural commodities will require improved productivity from land currently in cultivation as well as expansion into marginal land for agricultural use (1). In either case, one of the biggest problems facing agriculture is increased salinity in the soil solution. Salinity affects land that receives little rain ( Molecular mechanisms underlying regulation of cellular Na+ ion homeostasis during plant growth in salt Recently, much has been learned about signal transduction pathways that regulate plant ion homeostasis during salt stress (5). In a genetic screen designed to identify components of the cellular machinery that contribute to salt tolerance in Arabidopsis thaliana, three Salt-Overly-Sensitive genes (SOS1, SOS2, and SOS3) were found to function in a common pathway (6-9) [Figure 3 – SOS model]. Loss-of-function mutations in SOS1 render plants extremely For a complete picture of intracellular Na+ regulation during plant growth in salt, it will be necessary to understand To keep cytoplasmic levels of Na+ low, plant cells require the concerted activity of a number of transporters including the H+-pumps to provide the driving force for Na+ transport. Many of the transporters that would be required for the cellular response to excess Na+ have been identified in plants; however, little is know about their regulation. With the recent demonstration of SOS1 as a target of the SOS3-SOS2 pathway, we have the first insight into mechanisms underlying regulation of cellular Na+ homeostasis during salt stress. Current studies are focused on determining if the H+-pumps are also targets of the SOS pathway using in vitro, in vivo and in planta assays. As additional targets are identified, we will determine if there is coordinate regulation of pathway components during abiotic stress signaling. Genetic and biochemical studies have demonstrated that SOS1 (a plasma membrane Na+/H+ exchanger) is critical for maintenance of cellular Na+ homeostasis and salt tolerance in Arabidopsis. Analysis of the deduced amino acid sequence of the SOS1 protein has shown that it contains a long C-terminal tail that is likely to be localized in the cytoplasm. Recently, a regulatory function for the C-terminus of a SOS1-like protein (ApNhaP) from the cyanobacterium Aphanothece halophytica has been demonstrated (19). ApNhaP is an exchanger with novel ion specificity in that it displays both Na+ and Ca2+-coupled H+ transport (exchange). When the long C-terminus of ApNhaP was replaced with the C-terminus from a related transporter from Synechocystis with different ion specificity, the ion specificity of the Aphanothece protein was changed. This result indicates that the C-terminus of ApNhaP contains structural elements that are critical for ion-specific binding. We are determining the function of the C-terminus in SOS1 by asking if it plays a role in the regulation of SOS1 transport activity. To answer this question, we are determining the ion-specificity of SOS1 and the effects of altered forms of the SOS1 C-terminus on ion transport and salt tolerance in vivo and in planta. Identifying the genes, the biochemical mechanisms and the signaling pathways that mediate salt tolerance in a halophyte Mechanisms controlling stimulus-induced changes in intracellular calcium Calcium (Ca2+) has emerged as a critical component of many pathways in plants, underlying growth and development by linking perception of physiological and environmental cues to cellular responses. Because cellular Ca2+ levels are tightly regulated, small changes in cytosolic Ca2+ levels provide information for the modification of enzyme activity and gene expression needed for the subsequent responses. Stimulus-induced changes in intracellular Ca2+ are generated by transport proteins that allow the downhill flow of Ca2+ from a compartment in which the ion is present at high electrochemical potential (either outside the cell, or in the lumen of the vacuole or endoplasmic reticulum) into the cytoplasm in which Ca2+ is at lower potential. Numerous studies have implicated phosphoinositide-induced intracellular Ca2+ release (PICR) as part of the mechanism leading to stimulus-induced increases in intracellular Ca2+ levels in plants. Many of the components of the PICR pathway, including the intracellular messenger inositol 1,4,5-trisphosphate (InsP3), have been identified in plants as a result of genetic, biochemical and pharmacological studies. What is missing is the molecular identification of the InsP3 receptor (InsP3R), a Ca2+ channel that is responsible for these stimulus-induced increases in intracellular Ca2+. The recent identification of calcium binding proteins as novel protein ligands for the InsP3R in animal cells and the presence of plant calcium binding proteins with similar features and properties, provide an important new strategy to identify the receptor -- a component of the mechanism underlying Ca2+ regulation in plants. We are taking biochemical, molecular biological and genetic approaches to determine if the Arabidopsis CaBPs release Ca2+ from intracellular stores, if they interact with the InsP3R and to determine potential sites of CaBP/InsP3 interaction in the plant and in the cell. Identification of this receptor will be critical for understanding the specificity of Ca2+ signaling in plants and ultimately for designing strategies to modify the responses of plants to physiological and environmental cues.
1. O'Leary, J.W. 1995. Adaptive Components of salt tolerance. In: Handbook of plant and crop physiology. Mohammad Pessarakli, ed. Marcel Dekker, Inc, NY, pp. 577-585. 2. Rhoades, J.D. and Loveday, J. 1990. Salinity in irrigated agriculture. In: American Society of Civil Engineers, Irrigation of Agricultural Crops (Steward, B.A. and Nielsen, D.R., eds), Am. Soc. Agronomists, Monograph 30, 1089-1142. 3. Evans, L.T. 1998. Feeding the Ten Billion. Cambridge University Press, 247 p. 4. Penning de Vries, F.W.T. 2001. Food security? We are loosing ground fast! In: Nösberger J et al., eds. Crop Science: Progress and prospects, CABI Internatl Publ., pp. 1-14. 5. Zhu, J.-K. 2002. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247-273. 6. Wu, S.-J., Lei, D. and Zhu, J.-K. 1996. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8, 617-627. 7. Liu, J. and Zhu, J.-K. 1997. Proline accumulation and salt-stress-induced gene expression in a salt-hypersensitive mutant of Arabidopsis. Plant Physiol. 114, 591-596. 8. Zhu, J.-K., Liu, J. and Xiong, L. 1998. Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition, Plant Cell 10, 1181-1191. 9. Zhu, J.-K. 2000. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 124, 1-8. 10. Shi, H., Lee, B.-H., Wu, S.-J. and Zhu, J.-K. 2003. Over expression of a plasma membrane Na+/H+ antiporter improves salt tolerance in Arabidopsis. Nature Biotech. In press. 11. Halfter, U., Ishitani, M. and Zhu, J.-K. 2000. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA. 97, 3730-3734. 12. Shi, H., Ishitani, M., Wu, S.-J., Kim, C.-S. and Zhu, J.-K. 2000. A sodium-proton antiporter functions at the xylem/symplast boundary to control plant salt tolerance. Proc. Natl. Acad. Sci. USA. 97, 6896-6901. 13. Qiu, Q.-S., Guo, Y., Dietrich, M.A., Schumaker, K.S. and Zhu, J.-K. 2002. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99:8436-8441. 14. Apse, M.P., Aharon, G.S., Snedden, W.A. and Blumwald, E. 1999. Salt tolerance conferred by overexpression of a vacuolar Na+/H+-antiport in Arabidopsis. Science 285, 1256-1258. 15. Blumwald, E. 2000. Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 12, 431-434. 16. Zhang, H.-X. and Blumwald, E. 2001. Transgenic salt tolerant tomato plants accumulate salt in the foliage but not in the fruits. Nature Biotech. 19, 765-768. 17. Zhang, H.-X., Hodson, J., Williams, J.P. and Blumwald, E. 2001. Engineering salt-tolerant Brassica Plants: Characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc. Natl. Acad. Sci. USA 98, 12832-12836. 18. Qui, Q.-S., Guo, Y., Quintero, F.J., Pardo, J.M., Schumaker, K.S. and Zhu, J.-K. 2004. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the Salt-Overly-Sensitive (SOS) pathway. J. Biol. Chem. 279, 207-215. 19. Waditee, R., Hibino, T., Tanaka, Y., Tatsunosuke, N., Incharoensakdi, A. and Takabe, T. 2001. Halotolerant cyanobacterium Aphanothece halophytica contains a Na+/H+ antiporter, homologous to eukaryotic ones, with novel ion specificity affected by C-terminal tail. J. Biol. Chem. 276, 36931-36938. 20. Greenway, H. and Munns, R. 1980. Mechanisms of salt tolerance in nonhalophytes. Ann. Rev. Plant Physiol. 31, 149-190. 21. Zhu, J.-K. 2001. Plant salt tolerance. Trends Plant Sci. 6, 66-71. 22. Yang, J., McBride, S., Mak, D.-O.D., Vardi, N., Palczewski, K., Haeseleer, F. and Foskett, J.K. 2002. Identification of a family of calcium sensors as protein ligands of inositol trisphosphate receptor Ca2+ release channels. Proc. Natl. Acad. Sci. USA 99, 7711-7716. home | who we are | research | publications | tools | contact us |