metaFunction: A statistical tool in profiling functions in a
microbial community
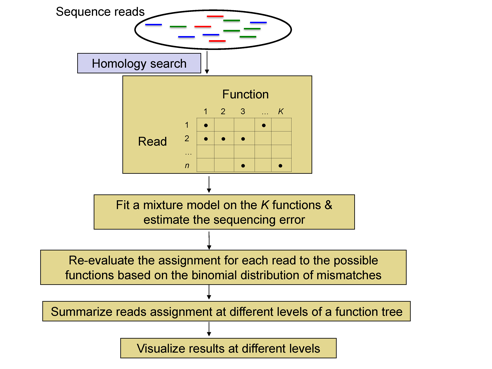
The overall workflow of metaFunction is shown
as below:
metaFunction package
can be downloaded here,
and the associated manual
file.
The
simulated sequence data (.fasta) can be found here.
R code for
analyzing a built-in example dataset:
library(metaFunction)
require(data.table)
require(mefa)
require(lattice)
data(ExampleData)
data(refseq)
data(subs)
pre=preprocess(ExampleData)
epsilon=0.01
est=Twosteps(pre, epsilon)
bootsize=100
boot.data=bootstrap(pre, bootsize, epsilon, method="B")
plot.abundance(boot.data)