TAEC: an R
package for Taxonomic Analysis by Elimination and Correction
TAEC, a new
homology-based approach for taxonomic analysis, utilizes the similarity in the
genomic sequence in addition to the result of an alignment tool.
This approach
consists of two main stages: the elimination stage and the correction stage. In
the elimination stage, the potential true genomes identified by removing false
genomes whose presence is most likely due to the presence of similar genomes in
a sample. In the correction stage, the abundances of the genomes remaining
after the elimination stage are corrected by utilizing the similarity between
genomes in a system of linear equations. The overall workflow of TAEC is shown
as below.
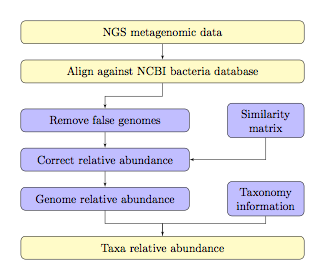
Note:
¾ The light yellow colored blocks
are implemented by a user and the light blue colored blocks are internally implemented by TAEC.
¾ The bacteria database could be replaced with virus
or other types of databases if needed.
¾ Similarity matrix is given for different lengths of
sequence reads, at 100pb, 250pb, 500pb, and 1000bp.
R package of TAEC can be downloaded: (Mac version)
and (linux version)
Note:
We have tested the TAEC package on R
version 2.14.1 (2011-12-22) and version 2.15.2 (2012-10-26) on Redhat and R version 3.0.2 (2013-09-25) on Ubuntu. No
errors associated with the different versions of R occurred. On the other hand,
we encountered errors associated with different versions of R on Mac OSX. In
order for the TAEC package to work properly on OSX, please upgrade your R to
the current version of 3.0.2 (2013-09-25).
Citation: Sohn M, An L, Pookhao N, Li Q. Accurate genome relative abundance
estimation for closely related species in a metagenomic
sample. BMC Bioinformatics 2014, 15:242 .